In 1997, an IBM developed computer program Deep Blue defeated world champion, Gary Kasparov, in chess. It was to show how artificial intelligence could evolve to beat its human master. Almost two years later, it was time for Google-owned DeepMind's AI program AlphaGo to defeat a human player in a board game called Go. Since then, AI has been touted as the game-changer that can solve problems by learning on its own. That has led scientists to apply AI in almost every field possible with varying results.
But now DeepMind has stunned the scientists once again by solving a 50-year-old "grand challenge", proving that AI is not far or has already taken over humans to some extent. Named AlphaFold, the AI program has managed to create a 3D model from the amino acid sequence that can help in drug development.
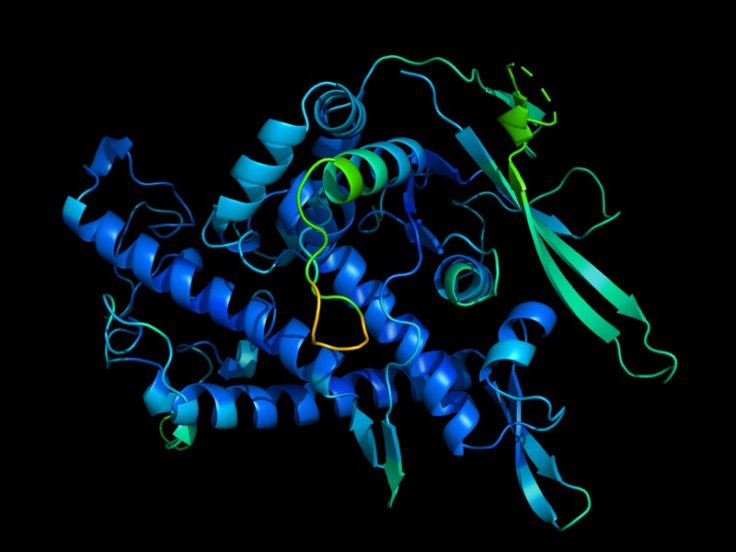
What is Protein Folding?
Thousands of proteins inside a cell keep it alive and are responsible for diseases. To assume its biological function, proteins fold or coil to form a 3D shape. The process from a linear shape of amino acids to curling is called protein folding. Hence, predicting the shape of the proteins will indicate the proteins' functions that play a major role in diseases including cancer.
For decades, scientists have struggled to create a 3D shape of protein folding that could aid in drug development to fight diseases. In 1994, scientists created a community experiment-cum-competition called Critical Assessment of protein Structure Prediction (CASP) to improve computational methods to predict protein structures. But until now, the experiment has had limited success. With AlphaFold, they have finally seen the problem being solved to some extent.
"This is a big deal. We have been stuck on this one problem – how do proteins fold up – for nearly 50 years. To see DeepMind produce a solution for this, having worked personally on this problem for so long and after so many stops and starts, wondering if we'd ever get there, is a very special moment," Professor John Moult, a computational biologist, Co-Founder and Chairman of CASP from the University of Maryland said.
How Has AlphaFold Evolved?
DeepMind's computer program participated in the computational biology experiment for the first time in 2018. By using the deep learning method in structural and genetic data, it was able to predict the distance between two amino acids in a protein. Then, it was able to produce a consensus model to predict how it would look.
However, it didn't work well. For the 2020 competition, DeepMind incorporated additional information in the AI network such as geometric and physical constraints. The aim was to predict the final structure of the protein sequence instead of predicting relationships between amino acids, John Jumper, DeepMind's project leader explained to the journal Nature. The AI model was trained with 170,000 protein structures and it worked.
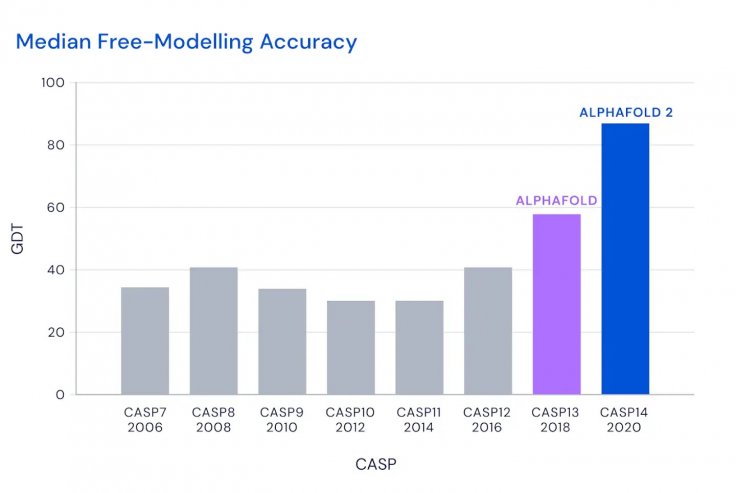
AlphaFold managed to score a median GDT of 92.4 and in the most challenging proteins, it managed a median of 87 that was 25-point higher than its closest competitor. Professor Venki Ramakrishnan, a Nobel laureate in chemistry and the President of the Royal Society called it a "stunning advance".
"This computational work represents a stunning advance on the protein-folding problem, a 50-year-old grand challenge in biology. It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research," he said.
While AlphaFold aced some challenges, it didn't do well in one protein, a mixture of 52 small segments that change each other's positions while assembling. Jumper told Sciencemag that the team would now train AlphaFold to solve those protein structures and others.









