University of Washington (UW) researchers found the missing genetic link behind the present-day Mongoloid race to two distinct pulses of Denisovans, a sister group to Neanderthals.
The research found that the mixing of modern humans with Neanderthals or Denisovans some 765,000 years ago resulted in the emergence of new East Asian populations and the disappearance of these two, who could have vanished as modern humans absorbed their lineage.
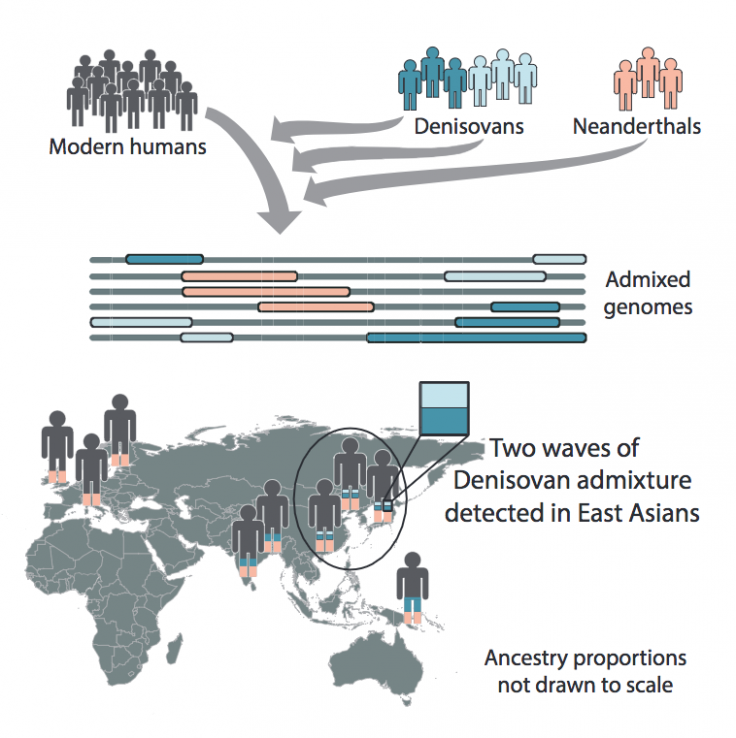
Lead author Sharon Browning, a professor of biostatistics in the UW School of Public Health, said: "Through DNA analysis, we knew there were at least two interbreeding events—with Neanderthals and with Denisovans. We now know there were at least three inbreeding events—at least one with Neanderthals, and at least two with Denisovans."
Anthropologists are aware that the genomes of certain populations, such as Papuan individuals, contained about 5% Denisovan ancestry, and that Denisovan ancestry throughout Asia was present to a lesser degree. However, comparison of DNA sequences shows a much closer Denisovan match among Han Chinese, Chinese Dai, and Japanese sequences, said Browning.
Denisovans are a distinct branch of the Homo family tree found in 2010 when a bone fragment and molars found in Denisova Cave in Siberia revealed a complete genome for a previously unknown human. But other than their DNA, no other trace about the Denisovans is there and anthropologists are still clueless about what they looked like, what they ate or what tools and technologies they possessed.

Browning, making use of a new reference-free statistical method Sprime (pronounced, s-prime) to existing genetic data from the 1000 Genomes project(link is external) and the Simons Genome Diversity Project(link is external), was able to search 5,639 whole genome sequences from present day people in Eurasia and Oceania and identify segments of DNA that were likely inherited from archaic human populations. The team compared genetic variants with Neanderthal and Denisovan sequences.
"We may be able to partially reconstruct the genomes of archaic human populations that did not leave remains with viable DNA for direct sequencing, such as populations that lived in warm and humid regions of the world where DNA decays rapidly," she said.









